SpinEvolution Driver
Problem
1) Table 1 of the SpinEvolution JMR paper compares the efficiency of SpinEvolution with that of SIMPSON in simulations of TPPM decoupling in CHn spin systems for a range of n values. In order to obtain data for this table, one may create an individual set of input files (main input file, coordinates, CS, and CSA files) for each n. This is not very convenient, however.
Make a single “universal” input file that can be used to run these simulations for any n. This file should load the coordinates from ch9.cor and CSA’s from ch9.csa. Hint: Use the extended molecule functionality and the -macro and -sys options.
2) The spin system size cannot be changed within a single simulation, so a separate computation for each n must be run even with the file you just created. This can be done manually, from a script, or from some other program. Do this from another SpinEvolution simulation. To be more specific: create an input file that runs such simulations for n from 1 to 6, loads the spectra obtained in these simulations, and saves them all in one file. Hints: Define n as a scan parameter and run the simulations for different n using the system() function. You will also need to use str(), save(), and load() functions.
Solution
1) tppmchn
****** The System *******
spectrometer(MHz) 500
spinning_freq(kHz) 15.625
channels C13 H1
nuclei *
atomic_coords ch9.cor
cs_isotropic 0.0 -0.3 0.5 0.5 0.2 0.2 0.2 0.0 0.0 0.0
csa_parameters ch9.csa
j_coupling *
quadrupole *
dip_switchboard *
csa_switchboard *
exchange_nuclei *
bond_len_nuclei *
bond_ang_nuclei *
tors_ang_nuclei *
groups_nuclei *
******* Pulse Sequence ******************************
CHN 1
timing(usec) (8)1024x1000
power(kHz) 0
phase(deg) 0
freq_offs(kHz) 0
CHN 2
timing(usec) (4 4)
power(kHz) 125 125
phase(deg) 0 15
freq_offs(kHz) 0 0
****** Variables **************************************
spinning_freq=15.625
n=8
t=500/spinning_freq/n
pulse_1_1_1=2*t
pulse_2_1_[1:2]=t
power_2_1_[1:2]=500/t
******* Options etc ***********************************
rho0 I1x
observables F1p
EulerAngles rep168
n_gamma 8
line_broaden(Hz) 2
zerofill *
FFT_dimensions 1 Hz
options -re -v0 -sysch{N}
********************************************************
-- This input file is run by the tppmx input file
-- The option -macro{N}=, where is the number of protons, must be specified from the command line
2) tppmx (the driver)
****** The System *******
spectrometer(MHz) *
spinning_freq(kHz) *
channels H1
nuclei H1
atomic_coords *
cs_isotropic *
csa_parameters *
j_coupling *
quadrupole *
dip_switchboard *
csa_switchboard *
exchange_nuclei *
bond_len_nuclei *
bond_ang_nuclei *
tors_ang_nuclei *
groups_nuclei *
******* Pulse Sequence ******************************
CHN 1
timing(usec) 0
power(kHz) 0
phase(deg) 0
freq_offs(kHz) 0
****** Variables **************************************
data=zeros(1024,7)
** The main loop: run tppmchn
scan_par n/1:6/
system("spinev tppmchn -macro{N}=" & str(n))
** Get the results from each loop cycle
data(:,1+n) = (load("tppmchn_re.dat"))(:,2)
** Get the labeling column (from the last cycle result)
data(:,1) = (load("tppmchn_re.dat"))(:,1), data_re
** Save the all results in one file
save(data,"tppmx_re.dat"), data_re
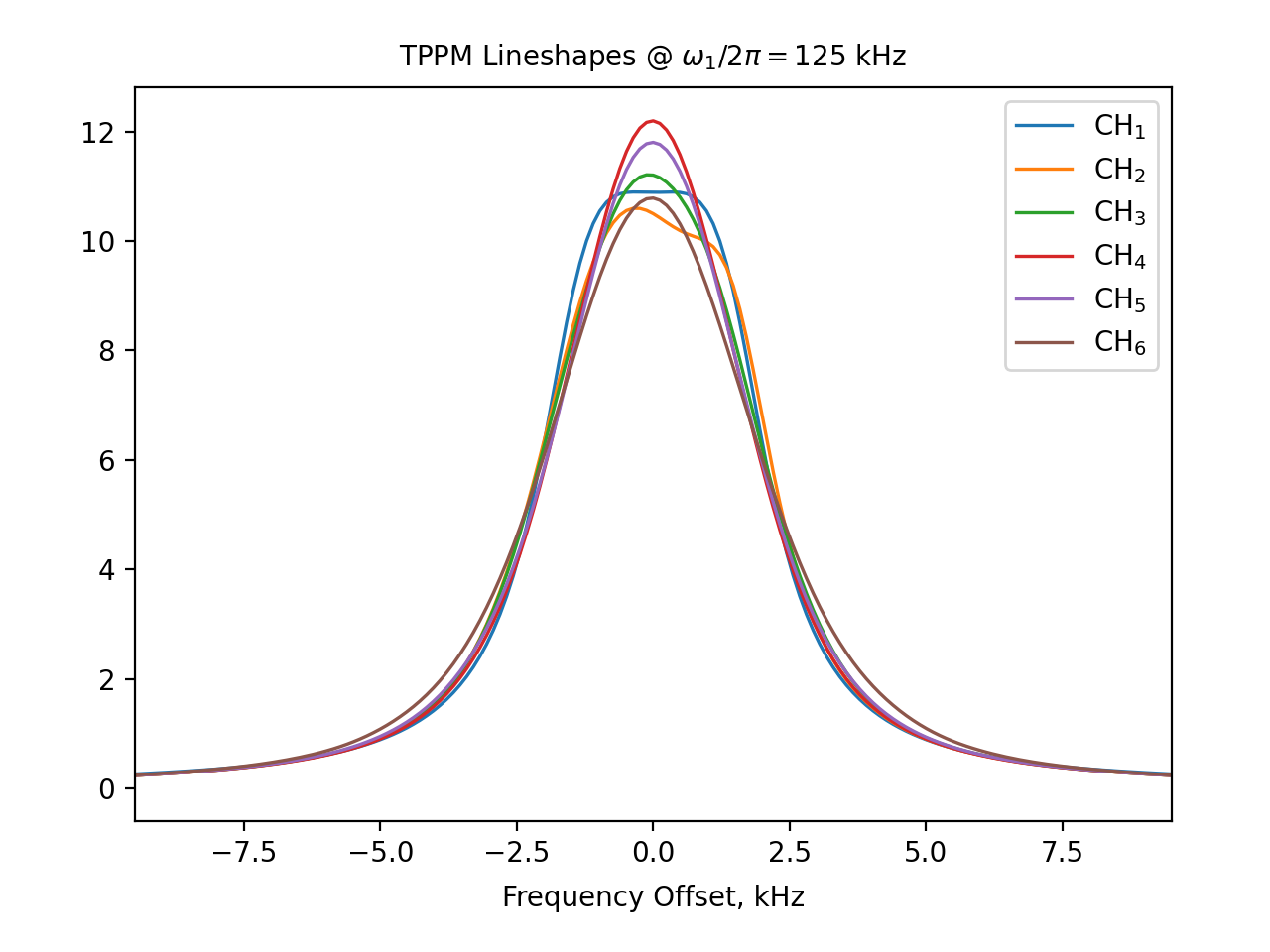
legend = "CH$_1$,CH$_2$,CH$_3$,CH$_4$,CH$_5$,CH$_6$"
fig_options=f"tppmx_re.dat --legend [legend] --xlim=-9.5,9.5 --xlabel 'Frequency Offset, kHz' --title 'TPPM Lineshapes @ $\omega_1/2\pi=125$ kHz'"
******* Options etc ***********************************
rho0 I1x
observables I1x
EulerAngles *
n_gamma *
line_broaden(Hz) *
zerofill *
FFT_dimensions *
options -o0 -split0 -py -pu
